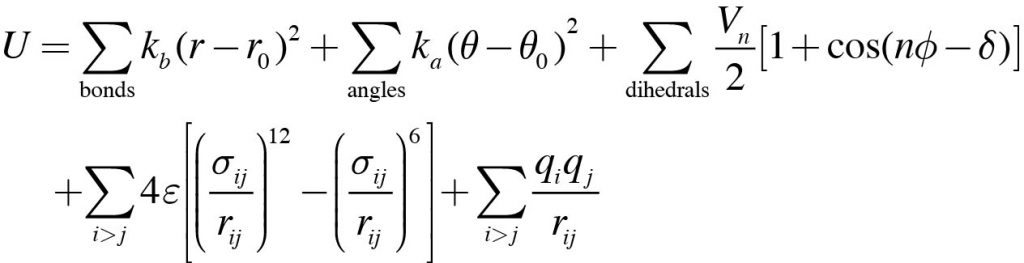Table 1 from CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field | Semantic Scholar

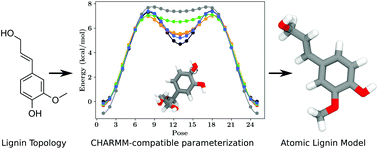
CHARMM general force field: A force field for drug‐like molecules compatible with the CHARMM all‐atom additive biological force fields - Vanommeslaeghe - 2010 - Journal of Computational Chemistry - Wiley Online Library

Exploiting the quantum mechanically derived force field for functional materials simulations | npj Computational Materials

BioExcel Webinar #58: CHARMM Force Field Development History, Features and Implementation in GROMACS - YouTube
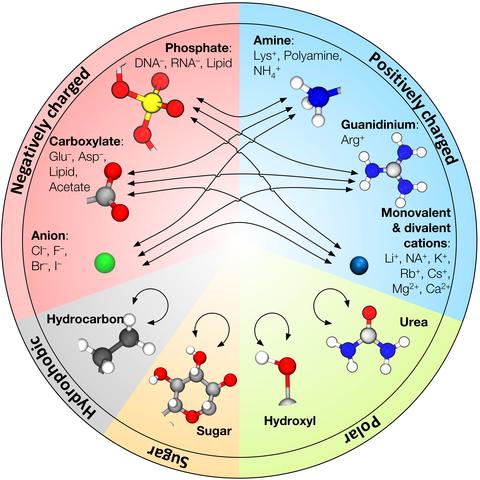
CUFIX: Non-bonded Fix (NBFIX) parameters for the CHARMM and AMBER force fields | The Aksimentiev Group
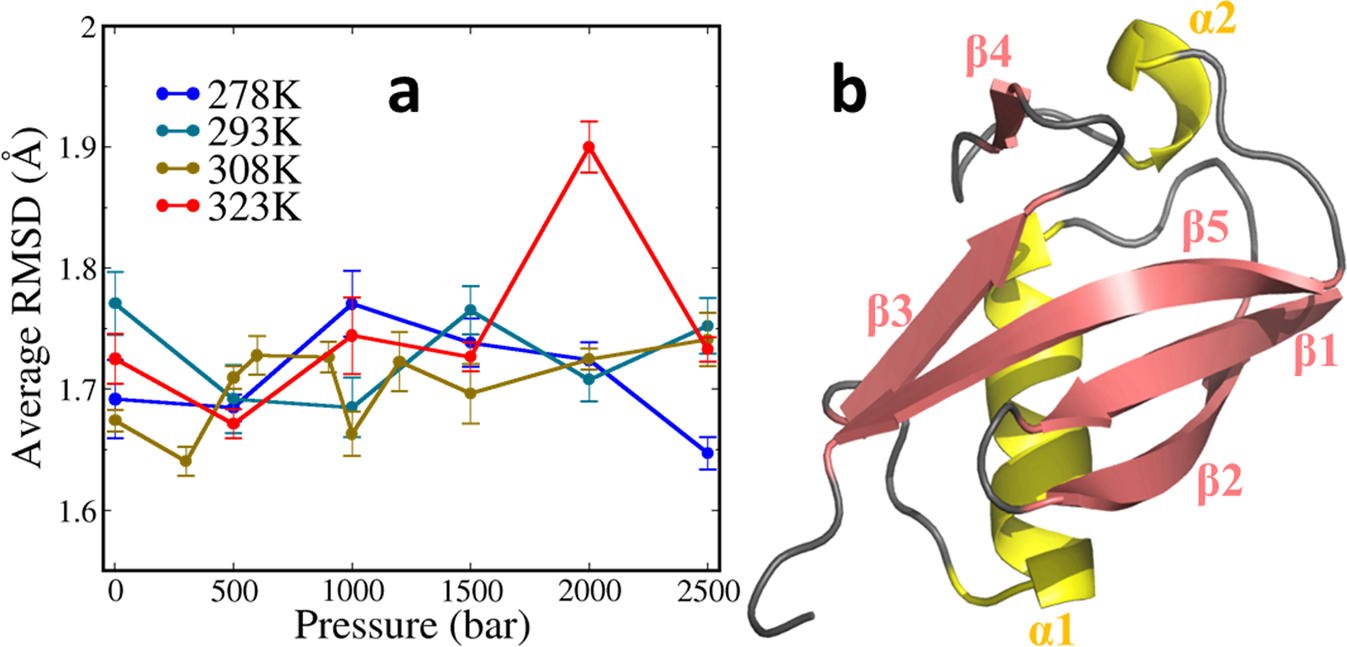
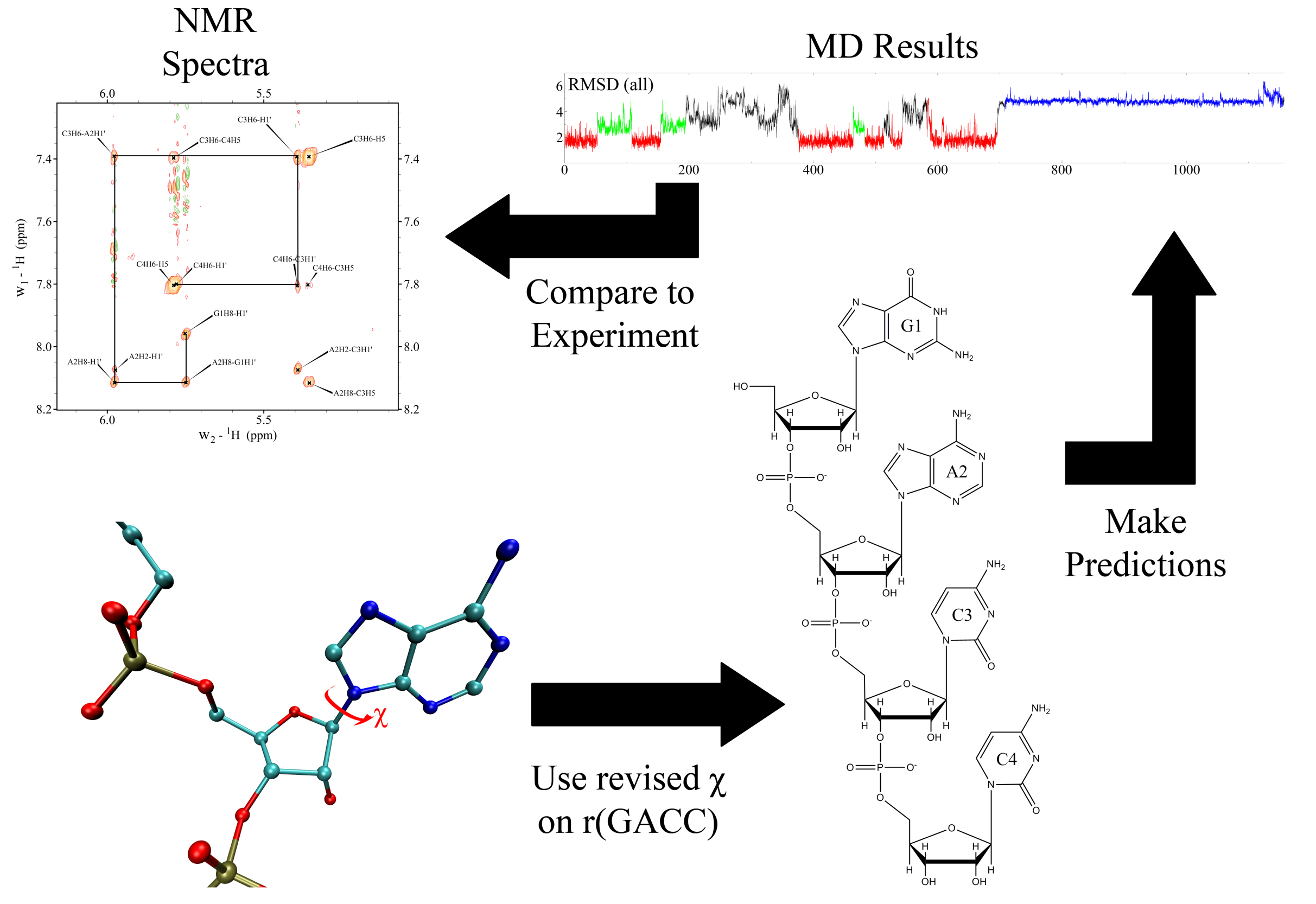
Validating the CHARMM36m protein force field with LJ-PME reveals altered hydrogen bonding dynamics under elevated pressures | Communications Chemistry

Non-bonded Fix Parameters For The Charmm And Amber - Md Simulation Force Field Transparent PNG - 480x390 - Free Download on NicePNG
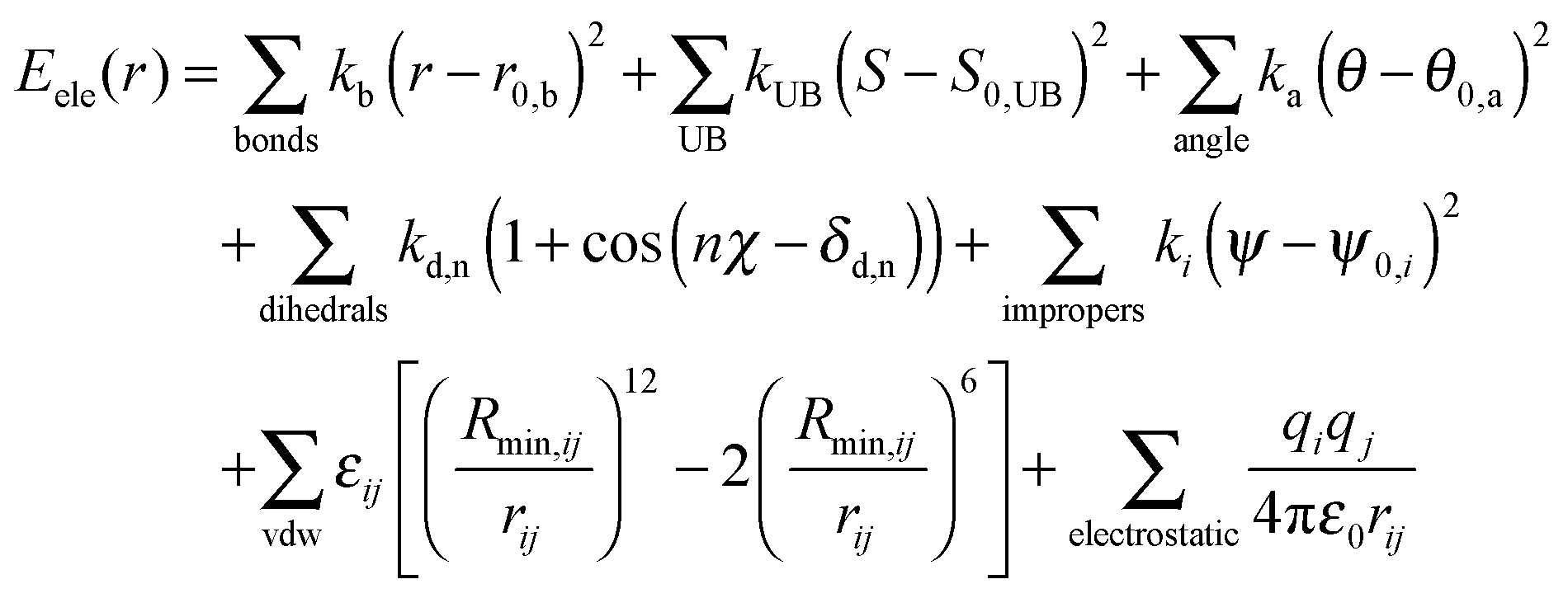
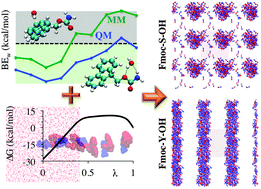
CHARMM force field parameterization protocol for self-assembling peptide amphiphiles: the Fmoc moiety - Physical Chemistry Chemical Physics (RSC Publishing) DOI:10.1039/C5CP06770G

Fig. S3. Optimization of an NBFIX correction of the CHARMM force field... | Download Scientific Diagram

CHARMM Force-Field Parameters for Morphine, Heroin, and Oliceridine, and Conformational Dynamics of Opioid Drugs | Journal of Chemical Information and Modeling

PMFF: Development of a Physics-Based Molecular Force Field for Protein Simulation and Ligand Docking | The Journal of Physical Chemistry B

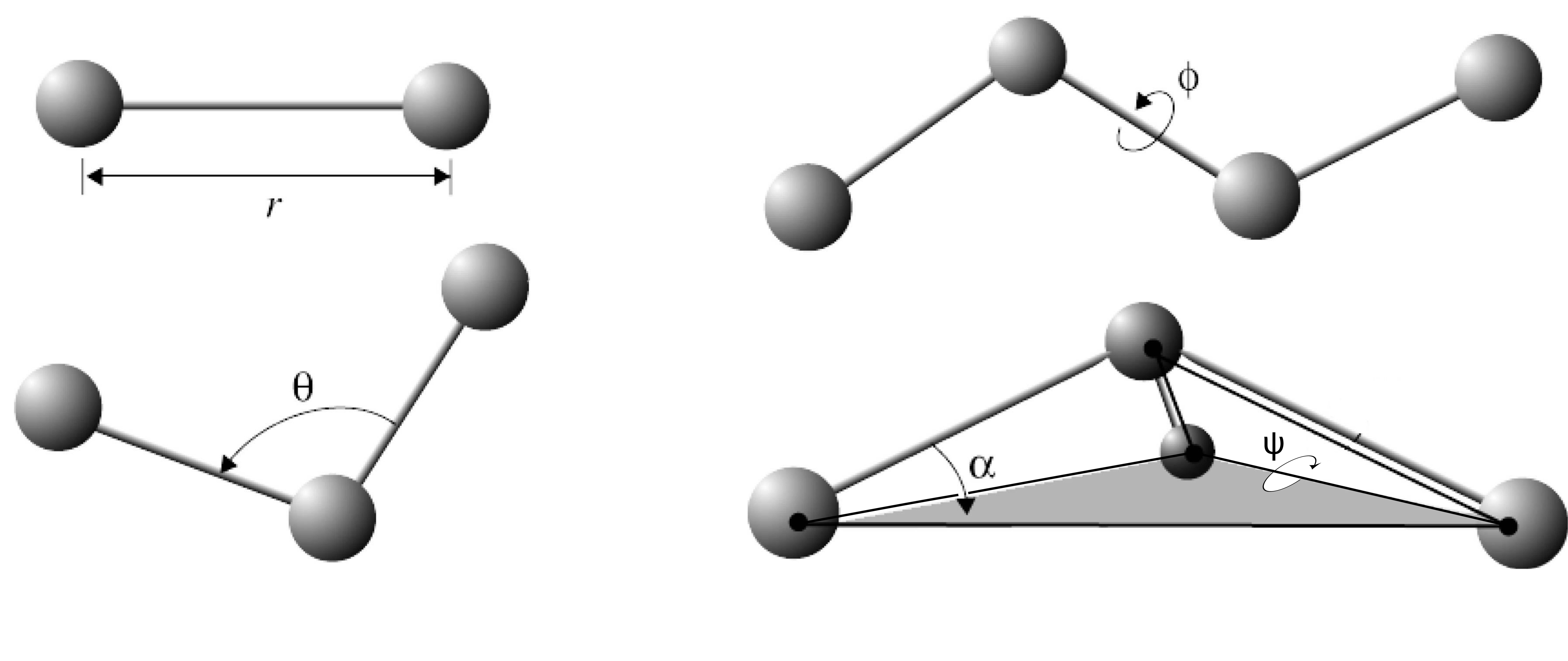
1: Schematic illustration of the bonded terms in the CHARMM force field... | Download Scientific Diagram